This note is a work-in-progress. We try to be as correct and fair as possible. If you find something wrong or have propositions for improvements, please open an issue in https://github.com/fluiddyn/transonic/issues.
Quick reminders¶
Transonic is a pure Python package (requiring Python >= 3.6) to easily accelerate modern Python-Numpy code with different accelerators (currently Cython, Pythran and Numba).
Pythran is an ahead-of-time compiler for scientific Python, with a focus on high-level numerical kernels, parallelism and vectorization.
Introduction¶
Transonic's documentation presents the project and covers a lot of technical details, but we think it is necessary to explicitly express the long-term vision behind the creation of Transonic. Transonic is a very young project (first commit in October 2018), with very few developers, actually currently (October 2019) only one core-developer (Pierre Augier) and 2 active contributors (Ashwin Vishnu and Pierre Blanc-fatin). We are of course very open to anyone interested in the project!
Status of the project¶
Transonic was originally built for packages of the FluidDyn project.

For some FluidDyn packages (FluidFFT, FluidSim, FluidImage), high performance is mandatory so we worked seriously on this aspect. We first used Cython and progressively switched to using only Pythran (in particular for its ability to strongly accelerate vectorized Python-Numpy code). Because Pythran (similar to Cython) optimized code on the module level and does not support Python classes (unlike Cython and Numba), our code had a "twisted" structure by the use of Pythran, with extra Pythran modules and functions that we wouldn't have with pure-Numpy code.
This motivated us to add a light runtime layer above Pythran to make it much more easy to use in packages, with a Python API similar to Numba's API. Several months of development followed (which are described here) and ultimately resulted in Transonic. With Transonic 0.4.0, we have gone beyond the original goals and it supports different Python accelerators (Cython, Pythran, Numba and possibly more in the future) as "backends". With Transonic,
Frontend (user API and code analyses) and backends (code generation and compilation) are well separated.
The different backends for different accelerators are written as classes with a simple and clear class hierarchy.
Users can use and switch between Ahead Of Time (AOT, with the
boostdecorator) and Just-In-Time (JIT, with thejitdecorator) compilation of functions / methods of classes. Thejitdecorator behaves like a cached JIT compilation for frameworks that only support AOT (Cython and Pythran). Additionally, blending compiled and interpreted Python code in a single module is now possible (unlike with pure Pythran).We have an API to describe types (and fused types). The code is built such that, it is simple to specify how these types have to be used and formatted for the different backends.
A lot of work has been done recently in the Python community to be able to annotate code with type hints. These hints are used in particular for documentation and for type checking (with tools like Mypy).
For Transonic, we also need type annotations but for a different purpose, namely compilation. One problem is that there is no standard way to describe Numpy arrays and "fused types" of Numpy arrays. This subject of type annotations for standard Python objects and for multi-dimensional arrays is actually very interesting and not so simple. For compilation, we need to be able to specify at least the possible dtypes and the possible numbers of dimensions, but also for some cases some fixed dimensions and the memory layout (C, Fortran, "C_or_F" and strided arrays). Unfortunately, as of now, there is no good and standard way to do that (tell us if we are wrong) so we had to invent and implement our own API.
Transonic code is seriously tested to check for example the stability of the generated Pythran, Numba and Cython codes. Tests are continuously run on Heptapod CI, Travis and Appveyor for Windows.
With this clean structure, adding a Transonic backend should be quite easy. Next time we write a backend (for example for Bohrium or Pyccel), we'll try to write it as a Transonic plugin and it should require very little modification in Transonic code.
To summarize, Transonic is a very young and small project. However, we think that we now have a clean base code (Disclaimer: the creator of Transonic is only a researcher in fluid mechanics with no experience in compiler development 😋 !).
A note on the current status of our Cython backend¶
Transonic 0.4.1 is already able to produce efficient Cython code
but more work is needed to reproduce advanced Cython features, like for example
with nogil:, C casting, from libc.math cimport ..., etc. See
our work on scikit-image numerical kernels.
A long-term vision associated with Transonic project¶
Currently, Transonic has very little known users. Of course, Transonic is used in FluidDyn packages. We also know few early adopters who now use Pythran through Transonic. In terms of package, pylandscape just started to use Transonic to be able to use Numba on Windows for a function for which Visual Studio does not manage to compile C++ Pythran code. We think that other packages already using Pythran or Numba could also benefit from using even the current version of Transonic (0.4.1), for example poliastro.
For most "simple" packages, the cost of the transition towards Transonic and the risks associated with this transition are small: (i) only small code modifications (usually going towards simpler code) and (ii) one more small pure-Python compile-time and run-time dependency. The bigger risk could be about Transonic no longer being maintained. However, even if something happens to the current maintainer, it should not be difficult to maintain Transonic usable and stable for a long time.
On the other hand, there are direct benefits and potential long-term opportunities: (i) cleaner code for packages using Pythran, (ii) Pythran / Numba power for packages not already using Pythran / Numba, (iii) easier benchmarks to test different accelerators, (iv) easy to use different accelerators for different OS and (v) choice between JIT or AOT compilations.
Then comes the question of the possible adoption of Transonic (or something similar to Transonic) in more used and/or fundamental packages of the ecosystem (QuTiP, Astropy, scikit-image, scikit-learn and even SciPy 🙂). Before addressing this subject, we think it is useful to present a global description of the current situation.
Current landscape of tools to accelerate numerical Python code¶
Overview¶
Python is a great language for teachers, scientists and data scientists. In terms of language, the design choices are strongly oriented towards readability, apparent simplicity, gentle learning curve and stability and not on pure performance. So let's stress that pure performance is not the only factor to consider! Readability, maintainability, generality of the code are also very important aspects!
With the Numpy API, Python is also very good to express numerical algorithms. With its huge scientific ecosystem, its a very efficient tool for fast prototyping of any kind of scientific libraries and applications. Since the number of people knowing Python is very large, projects using the Python language have a very large set of potential developers.
However, there are places in the code where performance really matters a lot, and Python is by default really bad for "number crunching".
In Python, all objects are "boxed". Even an integer (for example 1) is a box (a PyObject)
with inside a simple C 1. Passing the border between the Python world and the native world
therefore involves boxing or unboxing objects, which has a strong cost in terms of performance.
This partly explains why the loops in Python-Numpy as so inefficient. Therefore optimized
Python-Numpy code usually contains no or very few explicit loops.
Functions where the numbers are crunched are sometimes called "numerical kernels". Pure Python executed with CPython (the main Python implementation) is very bad for numerical kernels. Vectorized Numpy code is already much better, but one can still get much better performance with other languages or tools.
To avoid passing too much through the Python / native code border and to delegate expensive computations to optimized native code, one can create compiled functions with no or much less interactions with the Python interpreter.
PyPy¶

PyPy is an alternative Python interpreter. The slowest loops of pure Python code are accelerated via JIT compilation. However, Numpy is not written in pure Python and moreover it heavily relies on the CPython C API. PyPy now manages to emulate this API so it can run Numpy code but it has a strong cost in terms of performance. Therefore, it's unlikely that PyPy will ever be able to strongly accelerate numerical kernels written in Python-Numpy. (See also the very interesting long-term project HPy)
Note that the Transonic tests run fine with PyPy3.6 7.2.
Cython¶

The standard way to deal with heavy computational tasks in Python is to write C-extensions. The modern way to write C-extensions is to use Cython, which is a language (a superset of Python with many C features) and a compiler that transpiles Cython code into C or C++. Cython is used for two very different things: (1) binding of C APIs and (2) replacement of Python for CPU bounded problems.
Cython is widely used in all the scientific Python ecosystem. It is a very powerful language and a mature project. Great! However, Cython has also some issues:
Cython is currently unable to accelerate high-level Numpy code (except maybe when using Pythran in Cython).
To get something efficient, one need to write complicated code very different than standard Python-Numpy code. Cython is really a very powerful tool for experts who understand well Python, Numpy, C... and sometimes even the CPython C API.
There are much less potential developers able to write/understand good Cython code
Cython users lose the fast development without compilation which is one of the strength of Python.
Cython tends to encourage writing big C-extensions containing also Python functions/classes that do not need to be accelerated. It is often not useful for performance and tends to increase maintenance cost and to decrease development speed.
Low-level Cython codes are very specialized for CPU and it seems difficult to see how they could be used for GPU acceleration.
Cython also has a pure Python mode but (1) it's still very buggy and (2) Cython becomes a runtime dependency of the code.
Numba¶

Numba is a method-based JIT for Python-Numpy code, via transpilation into LLVM instructions.
This is a very similar technology than what Julia uses for its acceleration.
There is also a GPU backend that can be used together with Cupy.
Numba's user API is based on decorators which makes it very easy to use.
Note however that accelerating a vectorized code (optimized for execution with Numpy) necessitates
a lot of rewriting since Numba is not always able to strongly accelerate such Numpy code without explicit loops
(Pythran is usually better for high level code as shown by
these microbenchmarks).
Thus, only adding few jit decorators on a high level Numpy code may not lead to a
great speedup (Julia has actually similar problems, so it could be a fundamental problem
associated with the technology?).
Numba has many nice feature as jitclass, python mode, ufunc, gufunc, native functions, stencil, debug tools, etc...
A limitation of Numba is that it does not support proper ahead-of-time compilation allowing projects to distribute already compiled binaries.
Numba codes have a moderately big runtime dependency (the Numba package plus llvmlite, which is actually only 20 Mb big!).
Numba supports well the three main operating systems and many CPU architectures!
Numpy support is done through a reimplementation of NumPy in simple Python-Numpy!
This is interesting since it's then quite easy to contribute to this aspect of Numba. Note however, that it is not so simple to reimplement Numpy functions with only low-level Python-Numpy.
Pythran¶

Pythran is a ahead-of-time compiler for Python-Numpy codes, which are first optimized at the high level and then transpiled to very efficient (and highly templated) C++. The technology and the philosophy are very different than Cython and Numba.
First, Pythran compiles only pure Python-Numpy modules defining variables and functions
(see the supported modules and functions).
The only supplementary informations are instructions in Python comments
to say which functions have to be exported and for which input parameter types,
for example # pythran export my_great_function(int, float).
No fancy syntaxes unsupported by Python are required, as in Cython.
No need to give the types of local variables and returned objects.
Second, the resulting extensions never interact with the Python interpreter so that the GIL
is always released. So no need to use cython -a and to tweak the code to get rid of
the interactions with the Python interpreter as in Cython!
Other difference with Cython, Pythran uses SIMD through the C++ header library XSIMD. It means that advanced CPU instructions can be used even with high-level Python-Numpy code (i.e. code that can be executed in practice with only Numpy).
As a result of these technical characteristics, the performances of Pythran are in most cases impressive. It's usually not simple to beat Pythran with C++, Julia or Fortran (one has to be good and to work a bit). It is an unique tool since it can strongly accelerate both high-level and low-level Python-Numpy code.
From our experience of using Pythran and Numba, it appears that Pythran supports
more Python-Numpy-Scipy than Numba in no_python mode
(i.e. the mode for which you can get very good performance and for which the GIL can be released).
However, there is no "Python mode" in Pythran, so everything has to be implemented
in pure C++ without interaction with the Python interpreter.
Debugging and developing Pythran often require good skills in modern C++.
Note however than implementing Numpy or Scipy functions in Pythran is not so difficult (see for example this
PR on adding numpy.random distributions).
Since there is no interaction with the Python interpreter (you can just forget the GIL), Pythran is a great tool to parallelize code. For example, one can use Python threads to parallelize independent CPU bounded tasks. But OpenMP can also be used directly in Python-Numpy code! Of course, with all the C++ tools for parallelization and acceleration with GPU, Pythran has a strong potential in this area.
Pythran is much simple to use than Cython. There is (nearly) no special syntaxes to learn
(except the very simple mini-language for the signatures of the exported functions).
However, in practice, Python-Numpy codes have to be reorganized to use Pythran:
the numerical kernels have to be isolated from the rest of the code in special Pythran
modules containing only what can be compiled by Pythran and the Pythran
export commands with signatures. By offering an API similar to Numba,
Transonic completely solves this issue. Pythran itself does not support JIT mode,
but Transonic has a jit decorator that can be used with Pythran.
Note that it is also very easy to produce PyCapsule of native functions with Pythran.
Now, a few weaknesses of Pythran:
compilations can be lengthy and memory consuming
the Windows supports is not as good as the Linux/macOS support, partly because of some features of C++-11 seem not to be fully supported by Visual Studio! Note that Intel C++ compiler is also unable to compile (correct) C++-11 Pythran code!
Pythran is developed and maintained mainly by Serge Guelton. Even if there is a nice (and small) community of users and developers of the Pythran code, the bus factor is clearly equal to 1.
Many other projects¶
There are many packages to accelerate Python. Let's cite NumExpr, Cupy, Bohrium, Pyccel, Weld, JAX, PyTorch, uarray, statically, compyle, pystencil... Note that some of these projects have different approaches than just accelerating Python-Numpy code and rather provide their own API.
Overall comparison between Cython, Numba and Pythran¶
This table is taken from a presentation by Ralf Gommers (see also the video) on Scipy 1.0.
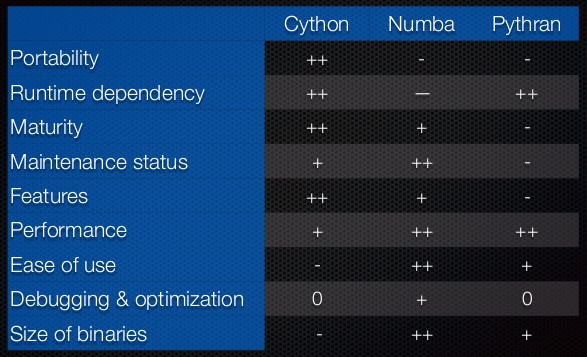
We interpret the symbols as very good (++), good (+), OK (0), bad (-)
and very bad (--).
We tend to think the comparison is a little bit unfair with Pythran. Pythran is no less mature compared to Numba. They are mature in different ways, and for some aspects, Pythran is even more mature than Numba.
Similarly, Pythran does not deserve a - for the features.
It would be very interesting to know which features are missing in Pythran
in the perspective of Scipy (maybe user-defined types?).
With the acceleration of high-level and low-level
Numpy codes, and automatic release of the GIL and OpenMP support,
we think that Pythran should deserve a + for the category "Features".
With these two changes, we can have a very different comparison.
Pythran and Numba still need some improvements but they appear as complementary tools
for the future of acceleration of Python-Numpy code.
Regarding Portability for Numba, it has recently
improved a lot
and the - seems too hard.
For Pythran, the portability is actually very good on Linux (and macOS)
since Pythran's C++ has been successfully tested on all architectures supported by Fedora.
Since recently, there was compilation problems on Windows because
Microsoft Visual C++ does not support well template forward declaration,
so the - seemed fair.
Fortunately, the next version of Pythran (that should be available on PyPI very soon)
will use by default clang-cl so that the whole Pythran test suite now also succeeds on Windows.
For the fun of it, we present an alternative table including also the solutions using Transonic
(with the boost decorator). The symbols in parentheses for Transonic correspond
to what could be obtained in few months if a little bit of work is invested into future
releases of Transonic.
| Cython | Numba | Pythran | Transonic-Cython | Transonic-Numba | Transonic-Pythran | |
|---|---|---|---|---|---|---|
| Portability | ++ | + | + | ++ | + | + |
| Runtime dependency | ++ | -- | ++ | + | -- | + |
| Ship independant binaries | ++ | -- | ++ | ++ | -- | ++ |
| Maturity | ++ | + | + | -- (+) | -- (+) | - (+) |
| Maintenance status | + | ++ | - | + | ++ | - |
| Features | ++ | + | + | - (+) | - (+) | 0 (+) |
| Performance | + | ++ | ++ | + | ++ | ++ |
| Ease of use | -- | ++ | + | + | ++ | ++ |
| Debugging & optimization | 0 | + | 0 | 0 | + | 0 |
| Size of binaries | -- | ++ | + | -- | ++ | + |
Transonic-Cython and Transonic-Pythran get a
+in "Runtime dependency" since the runtime dependencies (Transonic and its dependencies, i.e. Gast, Beniget, Astunparse and Black) are only small pure-Python packages, very simple to install anywhere just withpip(orconda).In terms of Maturity, we put
-- (+)for the solutions with Transonic because Transonic tasks are not so complicated (especially for the boost decorator) and it seems to me it could soon be very robust. The same argument also holds for the Maintenance status.For the Features, we also put
- (+)for the solutions with Transonic because many Cython and Numba features could soon be supported by Transonic.
We insert another more detailed table below. The symbol ✖️ is used for missing features.
| Cython | Numba | Pythran | Transonic-Cython | Transonic-Numba | Transonic-Pythran | |
|---|---|---|---|---|---|---|
| Modern Python style | -- | 0 | 0 | ++ | ++ | ++ |
| Python mode | ++ | ++ | -- | ++ | ++ | - |
| No-python mode | 0 | + | ++ | 0 | + | ++ |
| Numpy support no-python mode | -- | + | ++ | -- | + | ++ |
| Release the GIL | + | + | ++ | + | + | ++ |
| Fast math | ++ | ++ | ++ | + | ++ | ++ |
| SIMD loops | ++ | ++ | ++ | ++ | ++ | ++ |
| SIMD high-level | -- | -- | ++ | -- | -- | ++ |
| Parallel OpenMP | ++ | ++ | ++ | ++ | ++ | ++ |
| Native function (PyCapsule) | ++ | ++ | ++ | ✖️ (++) | ✖️ (++) | ✖️ (++) |
| User-def types | ++ | + | ✖️ (+) | ✖️ (+) | ✖️ (+) | ✖️ (+) |
| GPU support | -- | + | -- | -- | -- | -- |
| ufunc | + | + | ✖️ (+) | ✖️ | ✖️ | ✖️ |
| Stencil | ✖️ | + | ✖️ | ✖️ | ✖️ | ✖️ |
Micro-benchmark "high-level" and "low-level" implementations¶
Let's consider two implementations of a very simple function:
import numpy as np from transonic import jit @jit(native=True, xsimd=True) def fxfy(ft, fn, theta): sin_theta = np.sin(theta) cos_theta = np.cos(theta) fx = cos_theta * ft - sin_theta * fn fy = sin_theta * ft + cos_theta * fn return fx, fy @jit(native=True, xsimd=True) def fxfy_loops(ft, fn, theta): n0 = theta.size fx = np.empty_like(ft) fy = np.empty_like(fn) for index in range(n0): sin_theta = np.sin(theta[index]) cos_theta = np.cos(theta[index]) fx[index] = cos_theta * ft[index] - sin_theta * fn[index] fy[index] = sin_theta * ft[index] + cos_theta * fn[index] return fx, fy
fxfy is the high-level implementation that is simple and quite efficient with pure-Numpy.
I guess it is the implementation that most Python-Numpy users would write if they think about performance.
fxfy_loops is the implementation with explicit loops that can be strongly accelerated with Cython and Numba.
Note that the two implementations are not equivalent (the first one uses more memory)!
We reproduce here the result of a micro-benchmark using the transonic.jit decorator (smaller is faster):
fxfy (pure-Numpy) : 1.000 * norm
norm = 6.90e-04 s
fxfy_numba : 0.952 * norm
fxfy_loops_numba : 0.776 * norm
fxfy_pythran : 0.152 * norm
fxfy_loops_pythran : 0.784 * norm
Since Numpy is already quite efficient for the first implementation, the speed-up are only moderate. The faster result is obtained for the high-level implementation accelerated with Pythran, which runs 5 times faster than Numba with loops!
Analysis of the overall situation¶
Looking at this landscape, we see that there are many interesting projects but we also see a fragmented situation, with incompatible accelerators.
Numpy is quite efficient with high-level code but the two mainstream accelerators (Cython and Numba) are not good to accelerate this kind of code (see for example these microbenchmarks). So, after that the functions have been vectorized during the optimization for Numpy, they have to be devectorized to be accelerated with Cython and Numba. Then, this code with loops can't be used anymore with only Numpy. And it is often less readable and less general (only 1 number of dimensions). Having only accelerators efficient for low-level code is really sub-optimal.
Interestingly, it seems that we are in a local optimum for which Cython is good enough and the fundamental packages are reluctant to adopt other solutions, which therefore it inhibits progress. Cython is good for expert users and for the fundamental packages but not so good for less advanced users and smaller packages. The community is split and tools that could be used by "simple users" (simpler than Cython) are not (or less) used by advanced users. The investment in Python-Numpy accelerators is not very large. Even Cython has bugs that have to be avoided and no work in done to improve its capacity to accelerate standard high-level Python-Numpy code.
Pythran is clearly under-used and under-supported compared to its intrinsic quality and potential. We think the scientific Python community (and some of its leading heads) has to realize that Pythran is a great opportunity for our community. Pythran is one of the strongest link between the Python scientific community and the modern C++ community (see for example what is done with Xtensor). With Pythran, Python-Numpy can still shine in terms of performance for highly readable and high-level code, for example compared to Julia, for which low-level code and explicit loops are often needed to reach very high performance.
Some investment is put into Numba development, for example by Anaconda Inc. and Quansight. That's really great! But in the long-term, is it wise to only favor one technology (which has of course limitations)?
For users and developers of scientific packages (like FluidDyn packages), having this set of nice but fully incompatible accelerators is not nice. To stay in the standard and sure track used by the fundamental packages, we have to use Cython, which is not so well adapted for simple users and small packages. People have to learn Cython and develop/maintain Cython code. Moreover, the number of potential contributors/maintainers of these codes is much smaller than if they were in clean Python code. For example, students tend to know much less C and Cython than they know Python-Numpy.
People who want to try and benchmarks different solutions have to study them, and to reproduce nearly the same code for each accelerator, which is long, error prone and difficult to maintain. Refactoring a code by rewriting it is not a foolproof approach. For a new accelerator (for example, the Weld project), it's very difficult to be tried and used, which can slow down its potential adoption.
A loosely related problem is the future of computing with Python which involves different types of arrays (Numpy, Cupy, Dask, etc.). The accelerators won't be able to support all these arrays, so we'll need a kind of multiple dispatch mechanism for the numerical kernel. It could be done with Transonic (or something similar) at the user-defined function level.
The potential positive effects of a successful Transonic¶
Transonic could be one the elements that would allow a transition towards a better situation for developing high performance codes with Python. If Transonic (of something similar) starts to be adopted by enough packages, i.e. becomes a standard, this could have a good impact on the Python scientific ecosystem.
It would become extremely simple to accelerate any Python-Numpy code (also high-level code).
Accelerators would care about their ability to accelerate Python-Numpy code (focus on Numpy API) and would care about their Transonic backend, so that any packages using Transonic can easily enjoy potential improvements of one accelerator.
Fair competition between accelerators.
Tendency to unify the accelerator APIs.
More users would help the accelerators to improve.
Could Transonic become a standard? Using Transonic in fundamental projects?¶
We explained why we think Transonic is a good idea for the scientific Python community. But to become a standard, Transonic would have to be used in fundamental and/or big projects.
A simple example¶
Let's first show an example of code from which Transonic is able to produce efficient Cython, Numba and Pythran codes.
This code can be compared to
this Cython file in Scikit-image
(the nogil is not yet supported in Transonic 0.4.2, but Numba and Pythran automatically release the GIL).
import numpy as np from transonic import boost, Array, const Au = Array[np.uint32, "1d", "C", "positive_indices"] A = Array[np.int32, "1d", "C", "positive_indices"] M = Array[np.int32, "1d", "C", "memview"] @boost(boundscheck=False) def reconstruction_loop( ranks: Au, prev: A, next: A, strides: const(M), current_idx: np.intp, image_stride: np.intp, ): """The inner loop for reconstruction. ... """ current_rank: np.uint32 i: np.intp neighbor_idx: np.intp neighbor_rank: np.uint32 mask_rank: np.uint32 current_link: np.intp nprev: np.int32 nnext: np.int32 nstrides: np.intp = strides.shape[0] while current_idx != -1: if current_idx < image_stride: current_rank = ranks[current_idx] if current_rank == 0: break for i in range(nstrides): neighbor_idx = current_idx + strides[i] neighbor_rank = ranks[neighbor_idx] # Only propagate neighbors ranked below the current rank if neighbor_rank < current_rank: mask_rank = ranks[neighbor_idx + image_stride] # Only propagate neighbors ranked below the mask rank if neighbor_rank < mask_rank: # Raise the neighbor to the mask rank if # the mask ranked below the current rank if mask_rank < current_rank: current_link = neighbor_idx + image_stride ranks[neighbor_idx] = mask_rank else: current_link = current_idx ranks[neighbor_idx] = current_rank # unlink the neighbor nprev = prev[neighbor_idx] nnext = next[neighbor_idx] next[nprev] = nnext if nnext != -1: prev[nnext] = nprev # link to the neighbor after the current link nnext = next[current_link] next[neighbor_idx] = nnext prev[neighbor_idx] = current_link if nnext >= 0: prev[nnext] = neighbor_idx next[current_link] = neighbor_idx current_idx = next[current_idx]
This code has been adapted from an optimized Cython code and there is no high-level Numpy. Therefore, the performances of the generated Cython, Numba and Pythran codes are very similar. The original Cython code is more complicated with no (or small) performance gain (especially compared to Pythran compiled with clang).
Costs, gains, threats and opportunities¶
We now try to list the costs, gains, threats and opportunities associated with gradually using Transonic in such projects in the ahead-of-time mode. We consider here a super Transonic, very robust and with its most important features implemented.
Costs¶
Gradual code modifications, from Cython to Transonic code (which can be automatically translated in efficient Cython).
Less tunning for one particular accelerator, in particular for Cython. Some features not accessible from Transonic, for example pointers and
from libc.stdlib cimport malloc, free.More testing needed (?).
A new compilation-time and runtime dependency (again, small and pure-Python).
Gains¶
Simpler and nicer code than with Cython.
Can use other backends than Cython in the situations when they work and they are more efficient.
Threats¶
Possible Transonic bugs. Testing has to be super serious. Note that it seems feasible to build a very robust Transonic, especially for the ahead-of-time mode.
Issue with Transonic maintenance. We have to secure this, in particular by increasing the bus factor! Again Transonic is not so difficult to maintain.
Opportunities¶
Easily enjoy new Transonic backends or improvements of the accelerators.
Compatibility with exotic arrays in accelerated functions (if Transonic takes care of that).
Conclusion¶
We saw that there are many good tools to strongly accelerate Python-Numpy codes. Cython, Numba and Pythran use very different technologies and have their own strengths and weaknesses. Unfortunately, the accelerators are mostly incompatible and the community is split between expert users (who can enjoy the full power of Cython) and simple developers. A technical comparison of the different accelerators shows that Cython, Pythran and Numba are complementary tools for the future of acceleration of Python-Numpy code.
We tried to clearly explain the ideas behind the creation of Transonic. We hope some people will find them interesting and can be motivated to try Transonic, give feedback (https://github.com/fluiddyn/transonic/issues or more privately pierre.augier(at)univ-grenoble-alpes.fr) and even contribute!
If you think that this project is interesting, very simple ways to help are (i) to add a star to the project page on Github and (ii) to fill this very short survey. Note that we'll need to be able to prove that Transonic could be useful for some nice packages, so it's very valuable for us to get informal "declarations of interest" from identified developers of identified packages! And if we are allowed to use these declarations in proposals, it's even more useful!
Comments